Protein back-translation and alignment, discovering hidden protein homologies
PATH is a tool that addresses the problem of finding distant protein homologies where the divergence is the result of frameshift mutations and substitutions.
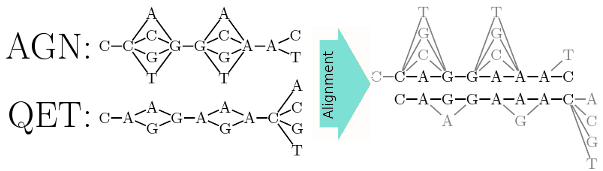
Given two input protein sequences, the method implicitly aligns all the possible pairs of DNA sequences that encode them, by manipulating memory-efficient graph representations of the complete set of putative DNA sequences for each protein.
The alignment algorithm finds two putative DNA sequences that have the best scoring alignment under an appropriate scoring system, designed to reflect the actual evolution process from a codon-oriented perspective. It incorporates a gap penalty that limits the number of frameshifts allowed in an alignment, to comply with the observed frequency of frameshifts in a coding sequence's evolution.
References
[1] Gîrdea, M. and Noé, L. and Kucherov, G.: Back-translation for discovering distant protein homologies, in Proceedings of WABI 2009, Philadelphia, September 12 – 13, 2009
[2] Gîrdea, M. and Noé, L. and Kucherov, G.: Back-translation for discovering distant protein homologies in the presence of frameshift mutations, Algorithms for Molecular Biology, Volume 5, January 2010