This page contains experimental datasets, results and associated scripts related to the JCB paper A Coverage Criterion for Spaced Seeds and Its Applications to Support Vector Machine String Kernels and k-Mer Distances. The content is divided into two parts that are related respectively to the sections 4.1. Coverage sensitivity and spaced seed string kernels and 4.2 Coverage sensitivity and alignment-free distance for sequence comparison of the paper.
The C++ is integrated into the Iedera 1.06α7 tool available at https://bioinfo.univ-lille.fr/yass/iedera.php
The Matlab (Octave compatible) code is available upon request to Donald E. K. Martin.
RFAM v11.0 →
(splitfasta.pl) →
Rfam/RF*.fa →
(main.pl) →
results
The initial dataset used is a multi-fasta file composed of 383004 sequences belonging to 2208 families from RFAM v11.0 [ftp://ftp.ebi.ac.uk/pub/databases/Rfam/11.0/Rfam.fasta.gz]. This multi-fasta file is processed to generate a set of (name normalized) fasta files using a script [splitfasta.pl].
The "Rfam" directory containing the fasta files is then processed by a second script [main.pl].
For each of set of seed(s) [seeds.txt], this script does most of the work :
svm-multiclass package [dir|url],
[dir]),
[corr.pl], and plots them [main-w3-n1.gp|main-w3-n2.gp|main-w4-n1.gp|main-w4-n2.gp].
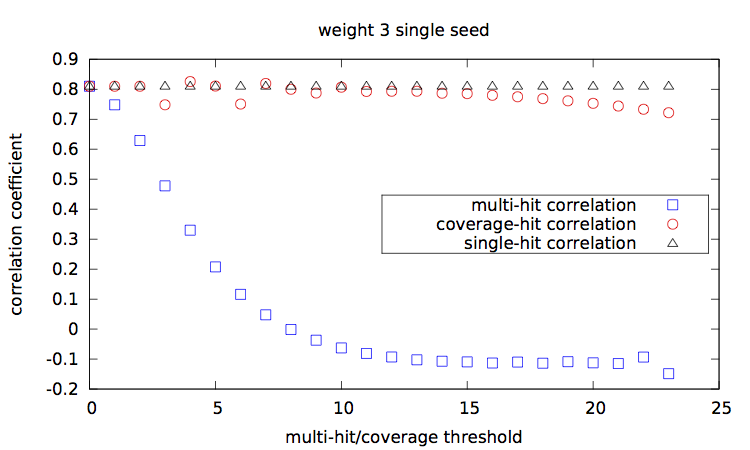
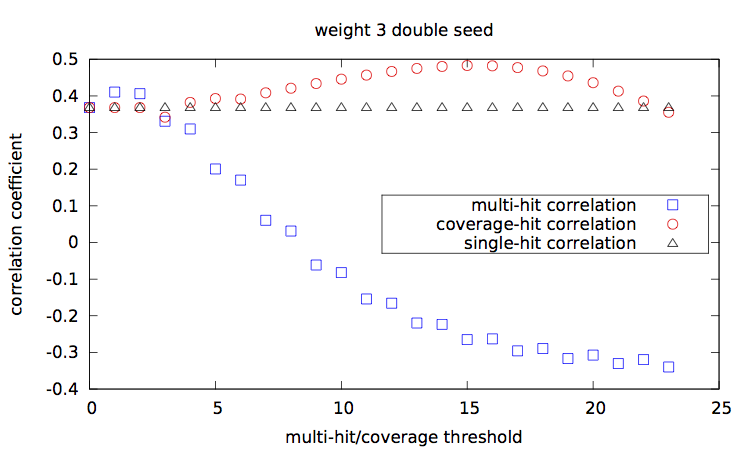
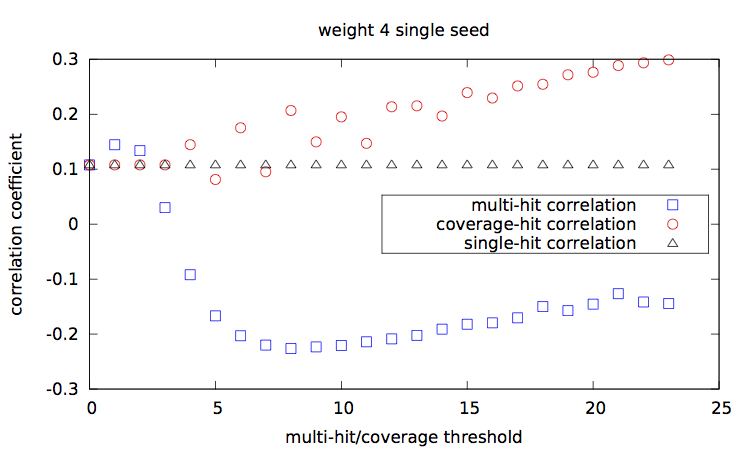
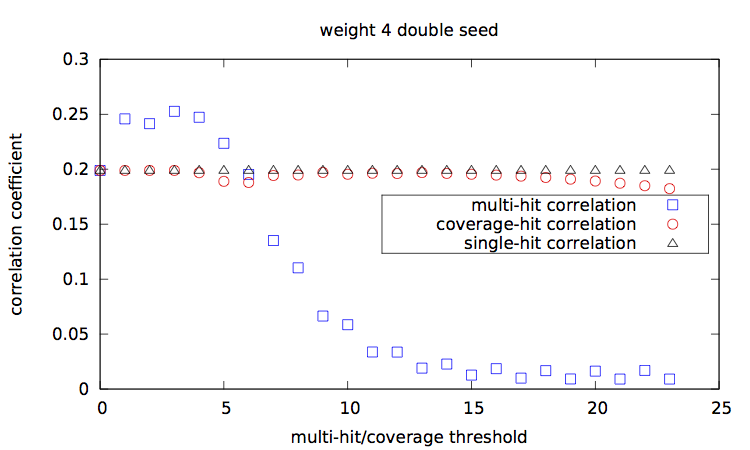
Given a set of seed(s), [seeds.txt], a main script [main.step.pl] computes, for each seed, for each alignment percentage of identity, and each of the 1000 trials, the Hamming distance, the estimated multi-hit and coverage values. During computation, it updates two correlation values, between the Hamming distances and each of the multi-hit values and coverage values. At the end, it outputs these two correlation values for each seed.
Correlation values [results.w3-n1.dat|results.w3-n2.dat|results.w4-n1.dat|results.w4-n2.dat] are then plotted with a script [results.gp].
[For additional tests, please see this webpage]